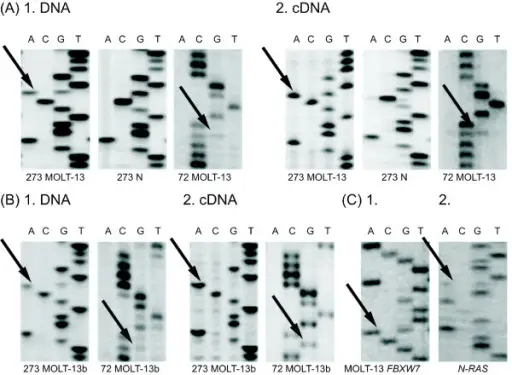
Mutations are alterations in the nucleotide sequence of the genome of an organism, virus, or extrachromosomal DNA.
Examples of mutations include:
- Point mutations
- Mutations in noncoding sequences
- Mutations in introns
- Mutations in exons
- Deletions
- Insertions
- Trinucleotide repeats
| Point mutations | A point mutation is alteration of a single base pair. The base substitution can be a silent mutation, which produces the same amino acid. The base substitution can be a missense mutation where the altered codon produces a different amino acid. The base substitution can be a nonsense mutation where a stop signal is produced. | – Point mutations are present in multiple tumor suppressor proteins cause cancer. Adenomatous Polyposis Col. Neurofibromatosis. Sickle-cell anemia |
| Mutations in noncoding sequences | Mutation in components of an organism’s DNA that do not actually code protein sequences. | – PTF1A enhancer mutation can cause Pancreatic agenesis.– RET enhancer mutation can cause Hirshsprung disease. |
| Mutations in introns | Mutations in intervening noncoding segments of DNA. | A splice site mutation produces the parathyroid hormone. |
| Mutations in exons | Mutation in part of a gene that will encode a part of the final RNA. | Many issues. |
| Deletions | Deletion is a mutation that results in loss of genetic material. Deletions can be small or large. | -DiGeorge syndrome – 22q11 deletion.-Cri-du-chat syndrome – microdeletion on the short arm of chromosome 5.-Williams syndrome – microdeletion of long arm of chromosome 7. |
| Insertions | Addition of one or more nucleotide base pairs into a DNA sequence. | Huntington’s diseaseFragile X syndrome |
| Trinucleotide repeats | Trinucleotide repeat expansions are a type of mutation where trinucleotide repeats in certain genes or introns exceed the normal, stable threshold. | – Fragile X syndrome – CGG repeats in fragile X mental retardation 1 (FMR1) gene.– Huntington’s disease – autosomal dominant trinucleotide CAG repeat expansion on chromosome 4. |