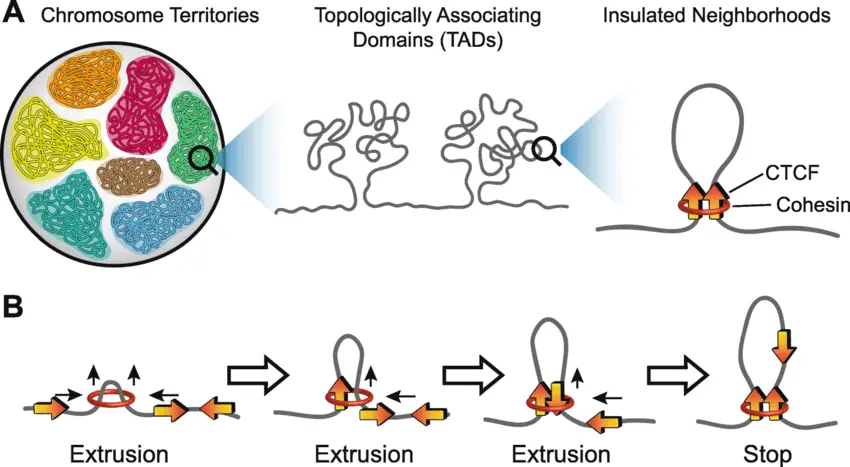
3D genome organization. a Each chromosome tends to occupy a particular region in the nucleus, defined as chromosome territories. Within a chromosome, there are regions with relatively high interaction frequencies, defined as topologically associating domains (TADs), and regions with relatively low interaction frequencies called TAD boundaries. Nested within each TAD are several sub-TAD domains, such as insulated neighborhoods, defined as DNA loops formed by CTCF homodimer (orange arrows), co-bound with cohesin (red ring), and containing at least one gene. b Extrusion/sliding model for TAD and sub-TAD loop formation: cohesin ring (red ring) facilitates the “sliding” of DNA through the ring structure to form a small loop. When bound CTCF (orange arrow) encounters cohesin, the DNA stops sliding on that side. The opposing side continues to slide through until a convergently oriented CTCF anchor motif is recognized and the insulator CTCF-CTCF–cohesin complex forms. Loops are less likely to form if two CTCF binding motifs are of tandem or divergent orientation
From association to mechanism incomplex disease genetics: the role of the 3D genome. Not altered. CC.
Yao Fu, Kandice L Tessneer, Chuang Li and Patrick M Gaffney
The genome is an organism’s complete set of genetic instructions. Each genome contains all of the information needed to build that organism and allow it to grow and develop.
There are ~3 billion DNA base pairs and 20 thousand protein-encoding genes.
Non-protein coding genetic sequences include:
- Enhancer regions
- Promoter regions
- Binding sites
- Non-coding regulatory RNAs
- Mobile genetic elements
- Telomeres
- Centromeres
DNA variation is commonly due to single nucleotide polymorphisms (SNPs) and copy number variations (CNVs).